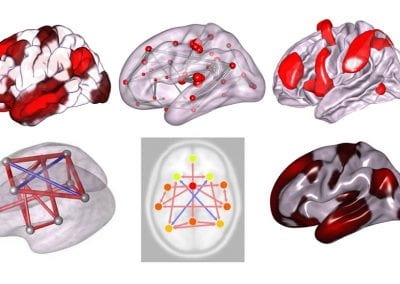
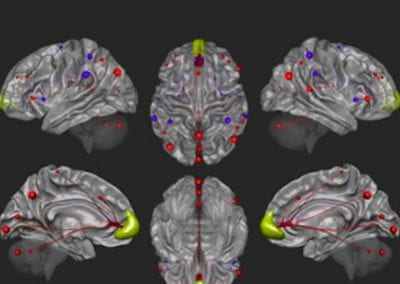
CONN Toolbox

CONN is a Matlab/SPM-based cross-platform open-source software for the computation, display, and analysis of functional connectivity Magnetic Resonance Imaging (fcMRI). CONN is used to analyze resting state data (rsfMRI) as well as task-related designs. Processing and analysis steps in CONN include:
1) Importing DICOM, ANALYZE, and NIfTI functional and anatomical files
2) Preprocessing of functional and anatomical volumes (e.g. SPM8/SPM12 realignment, slice-timing correction, outlier identification, coregistration, segmentation, normalization, smoothing)
3) Control of residual physiological and motion artifacts (e.g. scrubbing, aCompCor, ICA-based denoising, Global Regression)
4) Integrated quality control procedures and measures (e.g. FC histogram plots, BOLD signal carpet plots, Framewise Displacement, GCOR measures, FC-QC correlations)
5) Multiple connectivity analyses and measures, including Seed-Based Correlations (SBC), ROI-to-ROI analyses, complex-network analyses, generalized Psycho-Physiological Interaction models (gPPI), Independent Component Analyses (group-ICA), masked ICA, Amplitude of Low-Frequency Fluctuations (ALFF & fALFF), Intrinsic Connectivity (ICC), Local Homogeneity (LCOR), Global Correlations (GCOR), Multivoxel Pattern Analyses (MVPA), and dynamic connectivity analyses (dyn-ICA, sliding-window correlations)
6) Group- and population-level inferences and models, including ANOVA, regression, longitudinal, experimental, and mixed within- and between-subject designs. Control for multiple comparisons using parametric and non-parametric permutation/randomization techniques
When used within a distributed cluster, HPC, or multi-processor environment CONN can automatically parallelize most time-consuming steps in the fcMRI processing pipeline, allowing the analyses of hundreds of subjects in minimal time. CONN can be entirely controlled through a user-friendly GUI, or through batch scripts/commands if preferred.
The toolbox is developed in Matlab, and it is distributed both as Matlab source code and as a pre-compiled executable file (standalone release, no Matlab installations or licenses required).
For Installation and Manuals, please visit https://www.nitrc.org/projects/conn
Find about CONN Workshop: https://sites.google.com/view/conn/workshops
References
- Morfini, F., Whitfield-Gabrieli, S., & Nieto-Castanon, A. (2023). Functional Connectivity MRI Quality Control Procedures in CONN. Frontiers in Neuroscience, 17:1092125. doi: 10.3389/fnins.2023.1092125
-
Whitfield-Gabrieli, S. and Nieto-Castanon, A. (2012). Conn A Functional Connectivity Toolbox for Correlated and Anticorrelated Brain Networks Brain Connectivity, 2:125-41 (3418 citations, Consistently noted as being within the top 5 cited articles of the journal)
-
Chai X.J., Nieto-Castanon A., Ongur D., Whitfield-Gabrieli S. (2012) Anticorrelations in resting state networks without global signal regression NeuroImage, 59:1420-1428, (917 citations)
ART
ARtifact detection Tools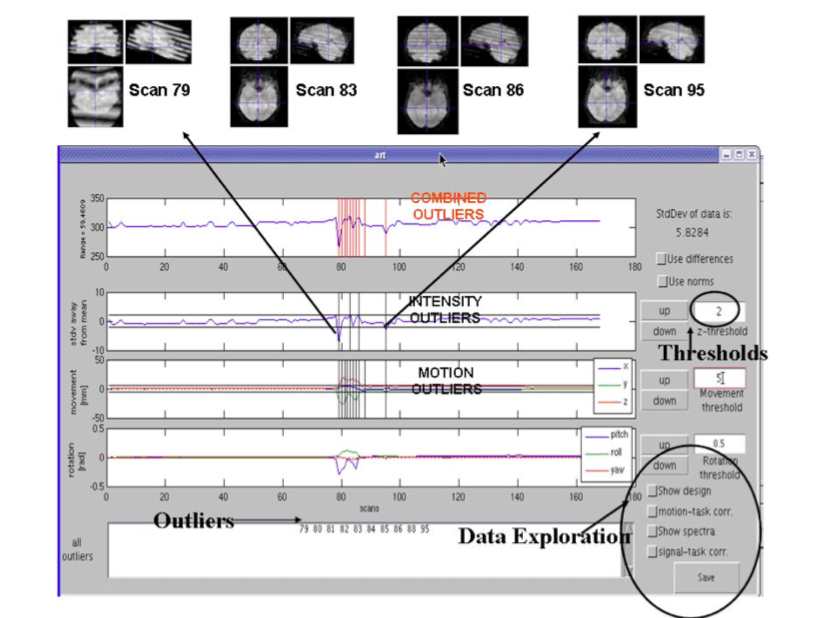
The ARifact detection Tools (ART) software package offers a set of tools that facilitate the identification of outlier values in fMRI time series that likely reflect artifacts and provides diagnostics tools which assist in appropriate design specification and model estimation. Specifically, it offers:
1) automatic and manual detection of global mean intensity and motion outliers in fMRI data that may be omitted from subsequent statistical analyses, interpolated or deweighted in the GLM;
2) methods for diagnosing the degree of stimulus-correlated motion in order to determine whether or not to include motion as confounds in the GLM;
3) methods for determining task correlation with global brain signal, in order to decide on global scaling options;
4) superposition of the experimental design on the time series for visual inspection; and
5) calculation of the power spectra of the experimental paradigm, as well as the motion, in order to allow users to select an appropriate high pass filter that does not filter out the frequencies associated with the task.
For Installation and Manuals, please visit https://www.nitrc.org/projects/artifact_detect
Reference
- Whitfield-Gabrieli S., Ghosh S., Nieto-Castanon A., Gollub R.L., Artifact Detection, Rejection and Quality Assurance of fMRI Data Increase Accuracy in Task Activation and Functional Connectivity Studies (In Revision).
REX
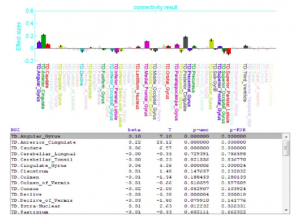
The rex tool is designed to extend those capabilities to permit the efficient extraction of image values and time series from single voxels, voxel clusters, and cluster collections. In addition to data extraction, the rex tool performs ROI- based analyses of functional data complementing SPM voxel-based analyses.
While rex is part of the larger BIT toolbox, it is a single MATLAB file that may be installed and used separately. It provides capabilities to extract image data from either a single image file or multiple volumes such as a time-series of images. The extraction volume can be either a single ROI or a collection of spatially disjoint ROIs. rex allows broad flexibility in the form of the extracted data, as it can return the values of a single voxel, a single cluster of voxels or a disjoint set of clusters. Various options are provided to generate descriptive statistics from clusters of voxels including mean, median, voxel-weighted mean, and one or more eigenvariates. To allow examination of experimental effects in units of percent signal change, the resulting extracted values can be scaled with respect to either the global brain mean, or the cluster time-series mean.
For installation and Manual, please visit https://www.nitrc.org/projects/rex/